

![]()
Accurate quantification of the number of amplifiable library molecules loading into a flow cell is one of the most critical steps in the next-generation sequencing (NGS) workflow in obtaining high-quality read data with NGS technologies. Loading insufficient amount of library DNA will result in low cluster density and reduced sequencing yield. An overabundance of library DNA may increase cluster density and result in poor quality data. Standard methods of NGS library quantification, by electrophoresis or spectrophotometry, have low sensitivity, are non-specific for adapter-bound DNA and typically require a large amount of library sample for analysis. With its greater sensitivity and broad dynamic range, qPCR is therefore seen as the gold standard for NGS library quantification, as it accurately measures the number of molecules that can serve as templates during library and cluster amplification, even with very dilute libraries.
Bioline has developed the JetSeq™ Library Quantification Kit, an optimized, robust SYBR® Green based qPCR kit that provides accurate quantification of Illumina based NGS libraries. The JetSeq Library Quantification Kit contains pre-diluted standards to minimize pipetting errors, a pre-qualified P5 and P7 Illumina adaptor sequence primer mix to ensure reproducible and precise qPCR results and an optimized buffer for dilution of NGS library samples.
![]()
Features
- Accurate - qPCR-based assay for quantification of only adapter-ligated library molecules, thereby enabling optimal flow cell loading for maximum data yield and quality
- Sensitive - reliable quantification of even low-yield libraries, ideal for both PCR and PCR-free library preparation methods
- Fast - delivers accurate assay results in as little as 90 minutes, thereby reducing time to results
- Convenient - contains a series of six pre-diluted DNA standards for rapid, simple standard curve development
- Economical - contains sufficient reagents and standards to quantify eighteen individual libraries on separate plates
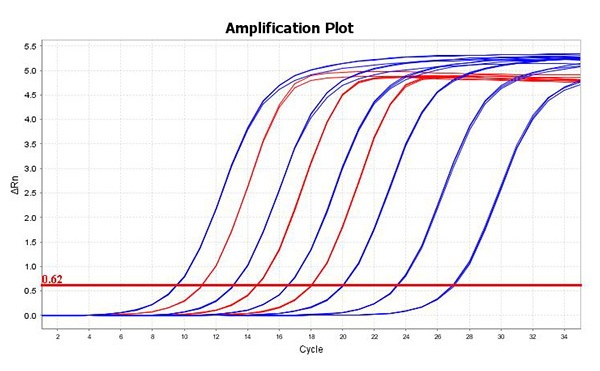
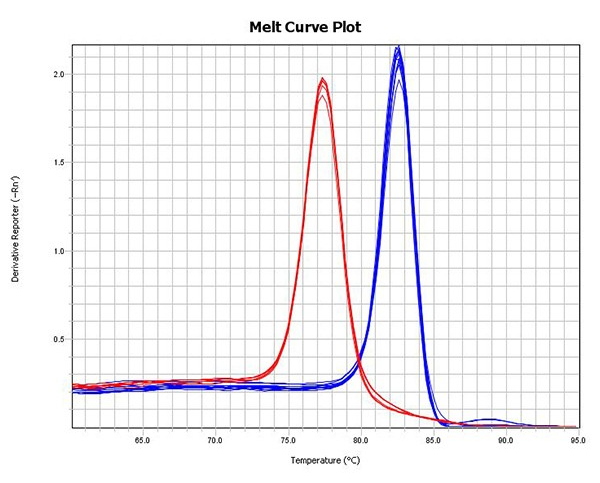
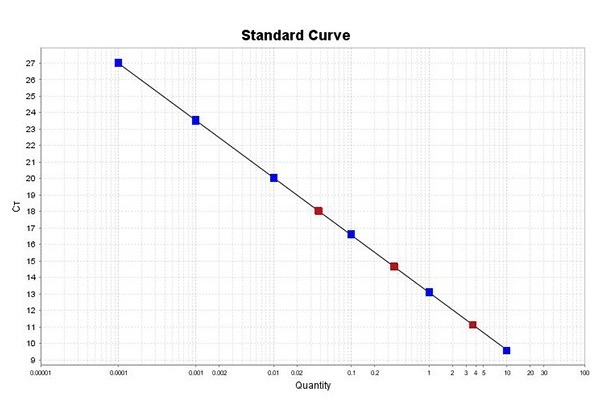
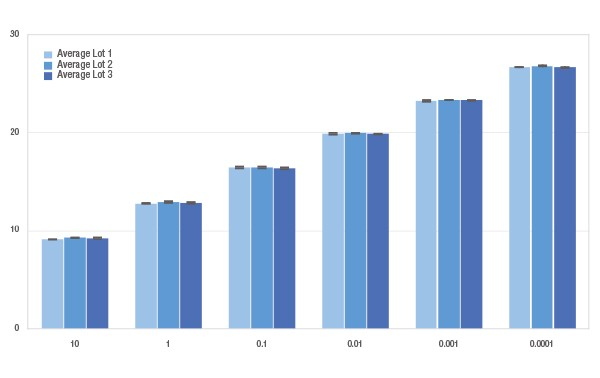
- Quantification of individual libraries or library pools prior to cluster amplification
- Quantification of libraries prior to pooling for multiplexed sequencing
- Quality control, optimization and troubleshooting of library construction processes or workflows



